Transposons families
Jump to navigation
Jump to search
| Prokaryotic elements |
|---|
|

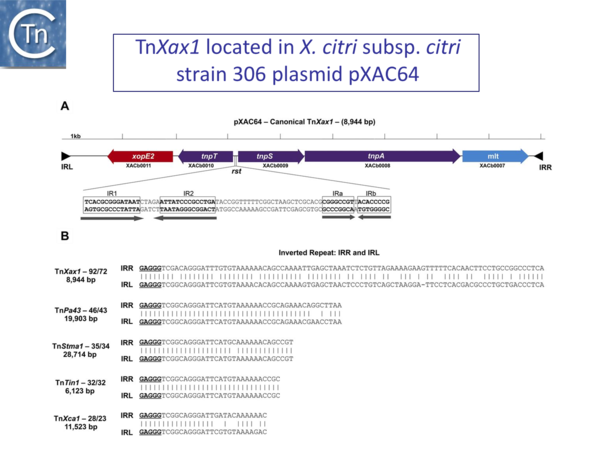
(A) Genetic organization of the canonical TnXax1 located in X. citri subsp. citri strain 306 plasmid pXAC64. Genes are indicated by colored boxes, with the direction of transcription shown by the arrowheads. Transposition-related genes are shown in purple, and passenger genes in red and blue. The presumed resolution site is located between the tnpT and tnpS genes, as observed in the transposable element Tn4651. This includes two palindromes: IR1 and IR2 are probably part of the core site at which recombination occurs, recognized by the TnpS recombinase, whereas IRa and IRb are potential binding sites for TnpT. The terminal inverted repeats (IRL and IRR) are shown as black triangles. The convention used for orientation of the transposon is that the transposase (TnpA) is transcribed from left to right. TnpA is closely related to other Tn3-like elements from Shewanella (ISSod9; 47% identity), Acinetobacter (ISAcsp1; 46%), Arthrobacter (ISArsp6; 43%), Azospirillum (ISAzs17; 41%), Salmonella (ISSwi1; 42%), and Nostoc (ISNpu13; 43%) species and to the mercury resistance transposon Tn5044 from X. campestris (84%). (B) IRs identified from other Tn3-like transposable elements. All include the GAGGG tips sharing sequence similarities to TnXax1 IRs. These are TnPa43 from Pseudomonas aeruginosa, TnStma1 from Stenotrophomonas maltophilia strain D457, TnTin1 from Thiomonas intermedia strain K12, and TnXca1 from X. campestris pv. vesicatoria plasmid pXCV183 (extracted from Ferreira et al., 2015).