General Information/IS History
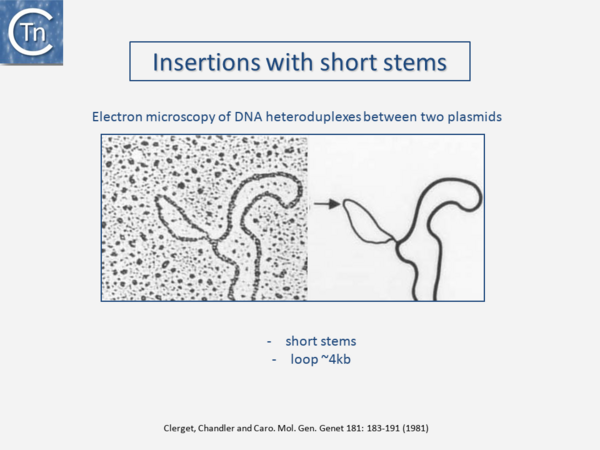
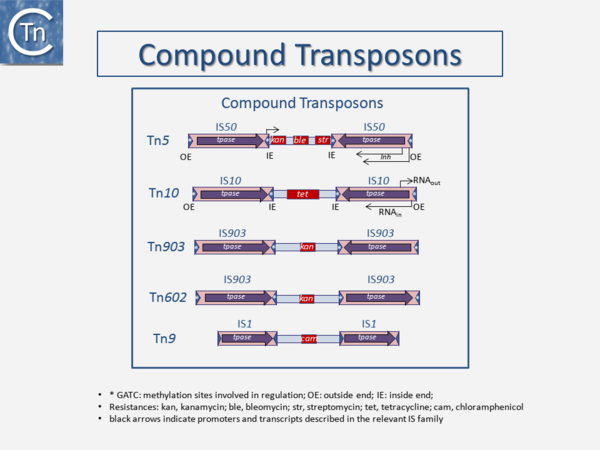
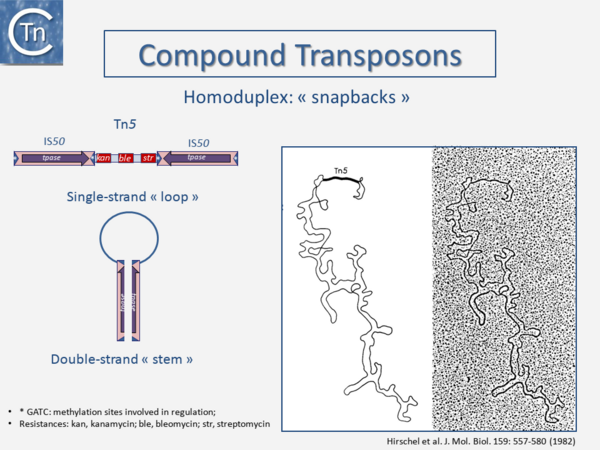
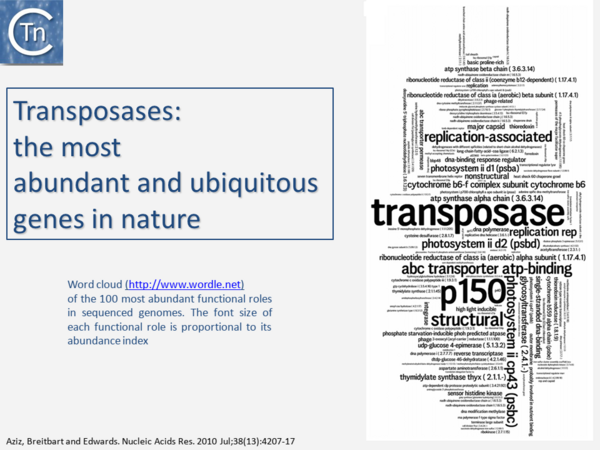
It is now over 40 years since the first IS were described. They were identified as short DNA segments found repeatedly associated with mutations in the gal operon and bacteriophage λ [1][2][3]. Shortly after, it was established that IS were normal residents of the Escherichia coli chromosome [4] sometimes present in multiple copies. They were shown to be involved in generating deletions [5] and in activating gene expression [6][7][6,7]. They were also identified as constituents of bacterial plasmids [8][9][10]. At about the same time, it was observed that antibiotic resistance genes could also be transferred or “transposed” from one plasmid to another [11][12][13] (Fig.1.2.1) and it was recognized that IS and “transposons” (a term originally coined by Hedges and Jacob [14]) were both members of a group of genetic entities: transposable or mobile genetic elements (TE or MGE). + Transposons and IS were identified by electron microscopy of heteroduplexes between plasmids or phage with and without the insertion or by homoduplexes (Fig.1.2.2) and (Fig.1.2.4) [15][16]) This relationship between IS and transposons was reinforced by the observation that different DNA segments carrying different genes could be translocated by two flanking IS (compound transposons) [17][18][19] (Fig.1.2.3) and (Fig.1.2.4). In many compound transposons one of the IS is inactivated or has reduced activity due to mutation. This tends to increase the coherence of the transposon in comparison to the autonomous activity of the individual IS. For example, in Tn5, the inside end (IE) of the left IS50 carries a mutation which both creates a better promoter sequence to drive expression of the resistance genes and introduces a UAA nonsense codon at the 3’ end of the transposase gene [20][21]. In the case of Tn10, the left IS10 differs from the functional right IS10 at a number of nucleotide positions in both in its regulatory region and in the transposase [22]. On the other hand, not all composite transposons possess an inactive copy of the flanking IS. In Tn9, flanked by two directly repeated copies of IS1, both IS are active [23]. It was also realized [24] that IS might be related to the controlling elements discovered by genetic analysis of maize several decades previously [25][26][27]. However, in spite of the observation that IS can be present in some bacterial species in extremely high copy numbers [28][29], little at the time prepared us for the subsequent recognition of the preponderant role they play in shaping genomes, of their extreme diversity and their widespread distribution (see reference [30](Fig.1.2.5).
Bibliography
- ↑ <pubmed>4567155</pubmed>
- ↑ <pubmed>4567154</pubmed>
- ↑ <pubmed>4555680</pubmed>
- ↑ <pubmed>4577900</pubmed>
- ↑ <pubmed>1101028</pubmed>
- ↑ <pubmed>5743437</pubmed>
- ↑ <pubmed>4610339</pubmed>
- ↑ <pubmed>1092668</pubmed>
- ↑ <pubmed>1092667</pubmed>
- ↑ <pubmed>1097422</pubmed>
- ↑ <pubmed>4612099</pubmed>
- ↑ <pubmed>1091625</pubmed>
- ↑ <pubmed>767328</pubmed>
- ↑ <pubmed>4609125</pubmed>
- ↑ <pubmed>6292435</pubmed>
- ↑ <pubmed>6281439</pubmed>
- ↑ <pubmed>385224</pubmed>
- ↑ <pubmed>368646</pubmed>
- ↑ <pubmed>1061090</pubmed>
- ↑ <pubmed>6244898</pubmed>
- ↑ <pubmed>6260374</pubmed>
- ↑ <pubmed> 6283536</pubmed>
- ↑ <pubmed>6313938</pubmed>
- ↑ <pubmed>339095</pubmed>
- ↑ <pubmed>17247459</pubmed>
- ↑ <pubmed>13433592</pubmed>
- ↑ <pubmed>15430309</pubmed>
- ↑ <pubmed>6265806</pubmed>
- ↑ <pubmed>6258088</pubmed>
- ↑ <pubmed>20215432</pubmed>