Difference between revisions of "IS Families"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
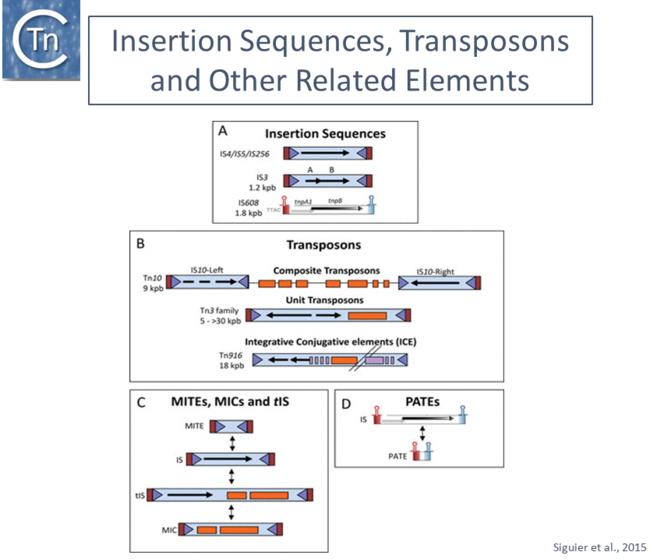
[[File:IS-web.png|thumb|650x650px|Organization of different insertion sequence (IS) -related derivatives. IS with DDE transposases (Tpases) and their derivatives are shown as blue boxes, '''T'''erminal '''I'''nverted '''R'''epeats as light blue triangles, and flanking direct target repeats as red boxes. The Tpase orfs are shown as black horizontal arrows. Passenger genes are shown as orange boxes and transfer functions (in the case of ICE) are shown as purple boxes. The single-strand IS are indicated with their left (red) and right (blue) subterminal secondary structures indicated. '''(A)''' IS organization. '''From top to bottom:''' a typical IS with a single Tpase orf; an IS in which the Tpase reading frame is distributed over two reading phases and requires frameshifting for expression; and the organization of a typical member of the single-strand IS family IS200/IS605. '''(B)''' Different IS-related TE. '''From top to bottom''': composite transposon Tn''10'' with inverted flanking copies of IS''10'' (note that the left IS''10'' copy is not autonomously transposable); a unit transposon of the Tn''3'' family; and an integrative conjugative element (ICE). '''(C)''' Relationship between IS, '''M'''iniature '''I'''nverted '''R'''epeat '''T'''ransposable '''E'''lements (MITE), '''t'''ransporter '''IS''' (tIS) and '''M'''obile '''I'''nsertion '''C'''assettes (MIC). '''(D)''' Generation of '''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements (PATE) from IS200/IS605 family members. Extracted from [https://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.MDNA3-0030-2014 Siguier et a., 2015.] |alt=]] | [[File:IS-web.png|thumb|650x650px|Organization of different insertion sequence (IS) -related derivatives. IS with DDE transposases (Tpases) and their derivatives are shown as blue boxes, '''T'''erminal '''I'''nverted '''R'''epeats as light blue triangles, and flanking direct target repeats as red boxes. The Tpase orfs are shown as black horizontal arrows. Passenger genes are shown as orange boxes and transfer functions (in the case of ICE) are shown as purple boxes. The single-strand IS are indicated with their left (red) and right (blue) subterminal secondary structures indicated. '''(A)''' IS organization. '''From top to bottom:''' a typical IS with a single Tpase orf; an IS in which the Tpase reading frame is distributed over two reading phases and requires frameshifting for expression; and the organization of a typical member of the single-strand IS family IS200/IS605. '''(B)''' Different IS-related TE. '''From top to bottom''': composite transposon Tn''10'' with inverted flanking copies of IS''10'' (note that the left IS''10'' copy is not autonomously transposable); a unit transposon of the Tn''3'' family; and an integrative conjugative element (ICE). '''(C)''' Relationship between IS, '''M'''iniature '''I'''nverted '''R'''epeat '''T'''ransposable '''E'''lements (MITE), '''t'''ransporter '''IS''' (tIS) and '''M'''obile '''I'''nsertion '''C'''assettes (MIC). '''(D)''' Generation of '''P'''alindrome-'''A'''ssociated '''T'''ransposable '''E'''lements (PATE) from IS200/IS605 family members. Extracted from [https://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.MDNA3-0030-2014 Siguier et a., 2015.] |alt=]] | ||
| − | {| class="wikitable" | + | {| class="wikitable" width="300pt" |
!++++ Prokaryotic elements ++++ | !++++ Prokaryotic elements ++++ | ||
|- | |- | ||
Revision as of 18:18, 1 June 2020

Organization of different insertion sequence (IS) -related derivatives. IS with DDE transposases (Tpases) and their derivatives are shown as blue boxes, Terminal Inverted Repeats as light blue triangles, and flanking direct target repeats as red boxes. The Tpase orfs are shown as black horizontal arrows. Passenger genes are shown as orange boxes and transfer functions (in the case of ICE) are shown as purple boxes. The single-strand IS are indicated with their left (red) and right (blue) subterminal secondary structures indicated. (A) IS organization. From top to bottom: a typical IS with a single Tpase orf; an IS in which the Tpase reading frame is distributed over two reading phases and requires frameshifting for expression; and the organization of a typical member of the single-strand IS family IS200/IS605. (B) Different IS-related TE. From top to bottom: composite transposon Tn10 with inverted flanking copies of IS10 (note that the left IS10 copy is not autonomously transposable); a unit transposon of the Tn3 family; and an integrative conjugative element (ICE). (C) Relationship between IS, Miniature Inverted Repeat Transposable Elements (MITE), transporter IS (tIS) and Mobile Insertion Cassettes (MIC). (D) Generation of Palindrome-Associated Transposable Elements (PATE) from IS200/IS605 family members. Extracted from Siguier et a., 2015.
| ++++ Prokaryotic elements ++++ |
|---|
|
| ++++ Eukaryotic elements ++++ |
|---|
|