Difference between revisions of "General Information/IS91 and ISCR"
| Line 3: | Line 3: | ||
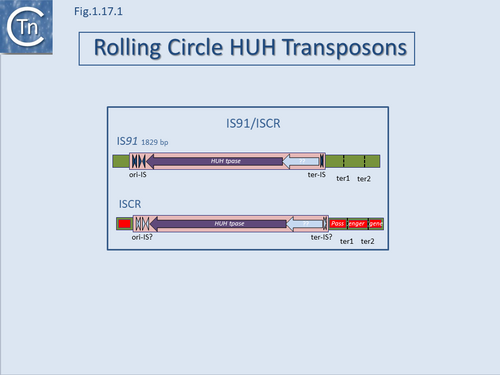
The transposons are shown as pink boxes; transposase as purple filled arrows; subterminal secondary structures essential for transposition are shown as filled dark blue (IS''91'') and pale blue (IS''CR'') arrow-heads. These, known as Ori-IS and ter-IS (in the case of ISCR this has not yet been clearly defined), are involved with initiation and termination of transposition on the rolling circle transposition model; larger, pale blue arrow is a Y-recombinase gene present in some IS''91'' family elements; secondary places which act as surrogate ends are also indicated to the right in the green flanking DNA; passenger genes are shown as red boxes. |alt=|border|center]] | The transposons are shown as pink boxes; transposase as purple filled arrows; subterminal secondary structures essential for transposition are shown as filled dark blue (IS''91'') and pale blue (IS''CR'') arrow-heads. These, known as Ori-IS and ter-IS (in the case of ISCR this has not yet been clearly defined), are involved with initiation and termination of transposition on the rolling circle transposition model; larger, pale blue arrow is a Y-recombinase gene present in some IS''91'' family elements; secondary places which act as surrogate ends are also indicated to the right in the green flanking DNA; passenger genes are shown as red boxes. |alt=|border|center]] | ||
| − | Although there has been no formal demonstration that these actually transpose, the CR is an orf which resembles the IS''91'' family Tpases<ref><nowiki><pubmed>23832240</pubmed></nowiki></ref>. The major feature of IS''CR'' elements is that they are associated with a diverse variety of antibiotic resistance genes and, particularly in the case of ''Pseudomonas'' IS''CR'', aromatic degradation pathways, both upstream and downstream of the Tpase ORF. | + | Although there has been no formal demonstration that these actually transpose, the CR is an orf which resembles the IS''91'' family Tpases<ref><nowiki><pubmed>23832240</pubmed></nowiki></ref>. The major feature of IS''CR'' elements is that they are associated with a diverse variety of antibiotic resistance genes and, particularly in the case of ''[[wikipedia:Pseudomonas|Pseudomonas]]'' IS''CR'', aromatic degradation pathways, both upstream and downstream of the Tpase ORF. |
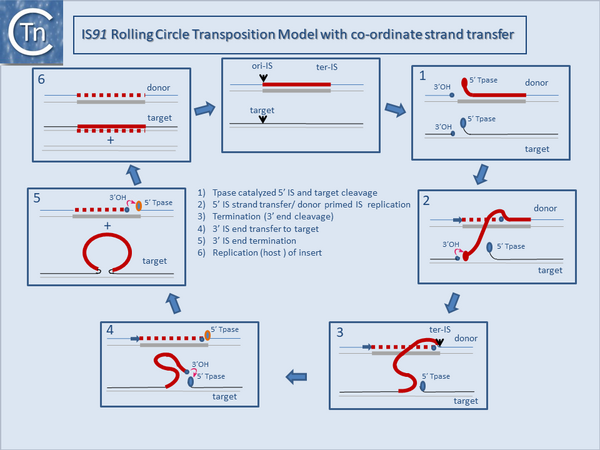
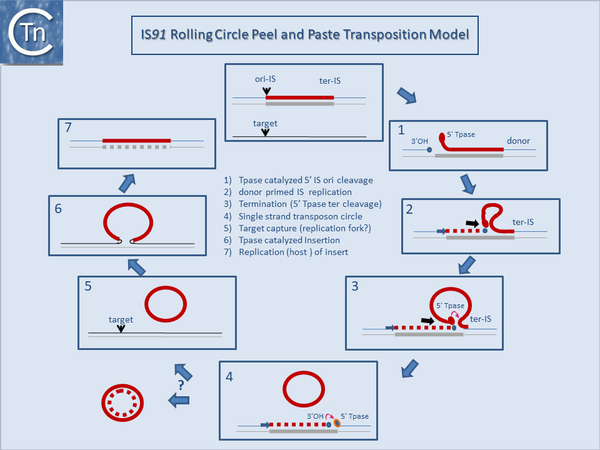
It is thought that these genes are transmitted during the rolling circle type of transposition mechanism postulated to occur in IS''91'' transposition [[:Image:1.17.2.png|(Fig.1.17.2)]]. This involves an initiation event at one IS end, polarized transfer of the IS strand into a target molecule and termination at the second end<ref><nowiki><pubmed>19709290</pubmed></nowiki></ref>. However, the [https://web.unican.es/ibbtec/en-us/about-ibbtec/team/members/member-detail?d=Fernando-de-la-Cruz-Calahorra-(IP) de la Cruz lab] has identified circular IS''91'' forms and it seems possible that these are transposition intermediates. An alternative model involving transposon circle formation is shown in [[:Image:1.17.3.png|Fig.1.17.3]]. This model is attractive since it would liberate the transposon circle intermediate to locating a target site following circle formation rather than requiring target engagement during the replicative transposition process [[:Image:1.17.2.png|(Fig.1.17.2)]]. | It is thought that these genes are transmitted during the rolling circle type of transposition mechanism postulated to occur in IS''91'' transposition [[:Image:1.17.2.png|(Fig.1.17.2)]]. This involves an initiation event at one IS end, polarized transfer of the IS strand into a target molecule and termination at the second end<ref><nowiki><pubmed>19709290</pubmed></nowiki></ref>. However, the [https://web.unican.es/ibbtec/en-us/about-ibbtec/team/members/member-detail?d=Fernando-de-la-Cruz-Calahorra-(IP) de la Cruz lab] has identified circular IS''91'' forms and it seems possible that these are transposition intermediates. An alternative model involving transposon circle formation is shown in [[:Image:1.17.3.png|Fig.1.17.3]]. This model is attractive since it would liberate the transposon circle intermediate to locating a target site following circle formation rather than requiring target engagement during the replicative transposition process [[:Image:1.17.2.png|(Fig.1.17.2)]]. | ||
Revision as of 18:53, 5 May 2020
A final example of the subtle line dividing IS and transposons is found in the IS91/ISCR group (Fig.1.17.1). IS91 was identified some time ago[1] and carries a single Tpase orf. More recently, a group of related elements, ISCR (IS with a "Common Region") was described [reviewed in [2][3]].
Although there has been no formal demonstration that these actually transpose, the CR is an orf which resembles the IS91 family Tpases[4]. The major feature of ISCR elements is that they are associated with a diverse variety of antibiotic resistance genes and, particularly in the case of Pseudomonas ISCR, aromatic degradation pathways, both upstream and downstream of the Tpase ORF.
It is thought that these genes are transmitted during the rolling circle type of transposition mechanism postulated to occur in IS91 transposition (Fig.1.17.2). This involves an initiation event at one IS end, polarized transfer of the IS strand into a target molecule and termination at the second end[5]. However, the de la Cruz lab has identified circular IS91 forms and it seems possible that these are transposition intermediates. An alternative model involving transposon circle formation is shown in Fig.1.17.3. This model is attractive since it would liberate the transposon circle intermediate to locating a target site following circle formation rather than requiring target engagement during the replicative transposition process (Fig.1.17.2).
Flanking gene acquisition is thought to occur when the termination mechanism fails and rolling circle transposition extends into neighboring DNA where it may encounter a second surrogate end. This type of mobile element may prove to play an important role in the assembly and transmission of multiple antibiotic resistance[6].