Difference between revisions of "IS Families/ISAzo13 family"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
| − | '''<big>T</big>'''his family, represented by 37 members in [https://isfinder.biotoul.fr/ ISfinder] emerged from the IS''NCY'' orphan group. It is based on both Tpase and '''IR''' sequence similarities [[:File:ISAzo13.1.png|(Fig.ISAzo13)]]. Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an HTH motif at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF ('''P'''rogrammed '''T'''ranscriptional '''F'''rameshifting) motif of 8 or 9 '''A''' while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)<ref | + | '''<big>T</big>'''his family, represented by 37 members in [https://isfinder.biotoul.fr/ ISfinder] emerged from the IS''NCY'' orphan group. It is based on both Tpase and '''IR''' sequence similarities [[:File:ISAzo13.1.png|(Fig.ISAzo13)]]. Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an HTH motif at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF ('''P'''rogrammed '''T'''ranscriptional '''F'''rameshifting) motif of 8 or 9 '''A''' while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)<ref><pubmed>16381877</pubmed></ref>. |
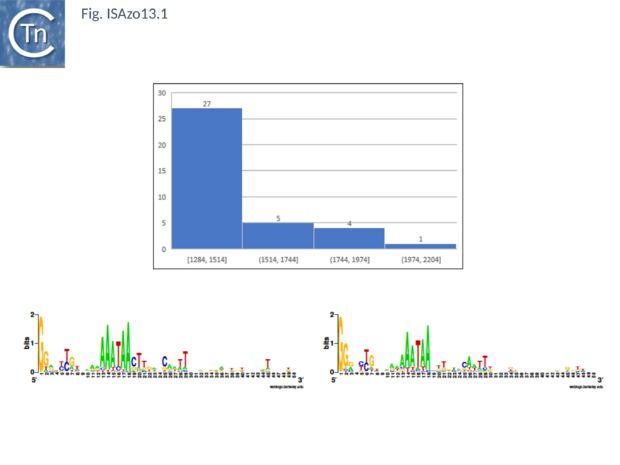
[[Image:ISAzo13.1.png|thumb|center|620x620px|'''Fig. ISAzo13.''' '''General [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] characteristics, average length and common ends. Top:''' Distribution of IS length (base pairs) of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] family members. The number of examples used in the sample is shown above each column. '''Bottom''': Left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format.|alt=]] | [[Image:ISAzo13.1.png|thumb|center|620x620px|'''Fig. ISAzo13.''' '''General [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] characteristics, average length and common ends. Top:''' Distribution of IS length (base pairs) of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISAzo13 IS''Azo13''] family members. The number of examples used in the sample is shown above each column. '''Bottom''': Left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format.|alt=]] | ||
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 23:57, 12 August 2021
This family, represented by 37 members in ISfinder emerged from the ISNCY orphan group. It is based on both Tpase and IR sequence similarities (Fig.ISAzo13). Insertion generates a 3bp AT-rich DR and the ends have a consensus GGa/g. Their Tpases are highly conserved with a probable DDE motif and an HTH motif at the N-terminus which could function as a DNA binding domain. Two members encode two orfs with a possible PRF (Programmed Transcriptional Frameshifting) motif of 8 or 9 A while the other members encode a unique orf which includes a triple lysine at the equivalent position (Gourbeyre, unpublished)[1].

Fig. ISAzo13. General ISAzo13 characteristics, average length and common ends. Top: Distribution of IS length (base pairs) of ISAzo13 family members. The number of examples used in the sample is shown above each column. Bottom: Left (IRL) and right IRR inverted terminal repeats are shown in WebLogo format.
Bibliography
- ↑ Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M . ISfinder: the reference centre for bacterial insertion sequences. - Nucleic Acids Res: 2006 Jan 1, 34(Database issue);D32-6 [PubMed:16381877] [DOI]