Difference between revisions of "IS Families/IS982 family"
| Line 4: | Line 4: | ||
====Organization==== | ====Organization==== | ||
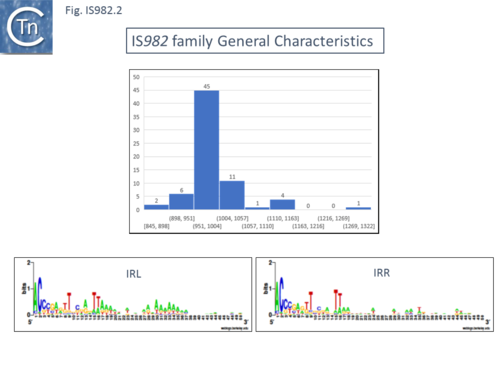
IS''982'' family members are between 962 and 1155 bp long [[:File:Fig. IS982.1.png|(Fig.IS''982'')]] and carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. IS''Sa4'' from ''[[wikipedia:Streptococcus_agalactiae|Streptococcus agalactiae]]'' generates 7 bp (or perhaps 9 bp) flanking DRs<ref><nowiki><pubmed>10607898</pubmed></nowiki></ref> and IS''Cce1'' from ''[[wikipedia:Clostridium_cellulovorans|Clostridium cellulovorans]]'' is flanked by 7 bp repeats<ref><nowiki><pubmed>10613891</pubmed></nowiki></ref>. | IS''982'' family members are between 962 and 1155 bp long [[:File:Fig. IS982.1.png|(Fig.IS''982'')]] and carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. IS''Sa4'' from ''[[wikipedia:Streptococcus_agalactiae|Streptococcus agalactiae]]'' generates 7 bp (or perhaps 9 bp) flanking DRs<ref><nowiki><pubmed>10607898</pubmed></nowiki></ref> and IS''Cce1'' from ''[[wikipedia:Clostridium_cellulovorans|Clostridium cellulovorans]]'' is flanked by 7 bp repeats<ref><nowiki><pubmed>10613891</pubmed></nowiki></ref>. | ||
| − | [[Image:Fig. IS982.1.png|thumb|center|500x500px|'''Fig. IS''982''.'''|alt=]] | + | [[Image:Fig. IS982.1.png|thumb|center|500x500px|'''Fig. IS''982''.''' The average length and common ends found in IS''982'' family.|alt=]] |
They encode a single orf of between 271 and 313 amino acids with a possible DDE motif but without a convincing conserved downstream K/R residue. They exhibit between 33% and 44% amino acid identity. Little is known about the transposition of these elements. | They encode a single orf of between 271 and 313 amino acids with a possible DDE motif but without a convincing conserved downstream K/R residue. They exhibit between 33% and 44% amino acid identity. Little is known about the transposition of these elements. | ||
Revision as of 14:03, 23 June 2020
General
The IS982 family has over 70 entries in ISfinder from over 40 bacterial and archaeal species. In the case of ISLpl4 from Lactobacillus plantarum identical copies have been detected in Leuconostoc mesenteroides, Oenococcus oeni, and Lactobacillus sakei indicating horizontal gene transfer. At least two members, IS982B[1] and IS1187 [see [2][3][4]] can provide a -35 hexamer in their right IR capable of forming a hybrid promoter with a resident -10 and activating neighboring genes.
Organization
IS982 family members are between 962 and 1155 bp long (Fig.IS982) and carry similar terminal IRs of between 18 and 35 bp with conserved ends: 5’-ACCC-3’ and generate DR of 6 to 8 bp e.g. ISSa4 from Streptococcus agalactiae generates 7 bp (or perhaps 9 bp) flanking DRs[5] and ISCce1 from Clostridium cellulovorans is flanked by 7 bp repeats[6].
They encode a single orf of between 271 and 313 amino acids with a possible DDE motif but without a convincing conserved downstream K/R residue. They exhibit between 33% and 44% amino acid identity. Little is known about the transposition of these elements.
Although the Tpase of a majority of members occupies a single reading phase, there are several examples in which the gene is distributed over two phases. It has been reported that a +1 nucleotide insertion in the Tpase orf of an ISLpl4 from Oenococcus oeni may undergo programmed translational frameshifting at a low rate[7]. Although this must be confirmed, it would represent the first functional case of +1 frameshifting in IS. The Tpase of archaeal element ISPfu3 is also distributed over two phases. ISPfu3 carries a potential transcriptional frameshift signal A7 (PTR) present in all five copies suggesting that ISPfu3 is active.